Lipid-anchored protein
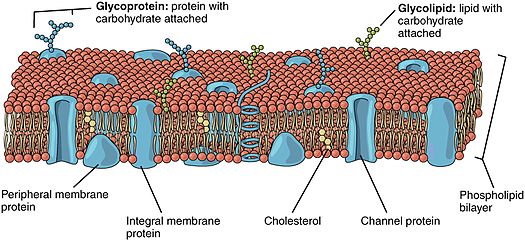
Lipid-anchored proteins (also known as lipid-linked proteins) are proteins located on the surface of the cell membrane [of what?] that are covalently attached to lipids embedded within the cell membrane. These proteins insert and assume a place in the bilayer structure of the membrane alongside the similar fatty acid tails. The lipid-anchored protein can be located on either side of the cell membrane. Thus, the lipid serves to anchor the protein to the cell membrane.[1][2] They are a type of proteolipids.
The lipid groups play a role in protein interaction and can contribute to the function of the protein to which it is attached.[2] Furthermore, the lipid serves as a mediator of membrane associations or as a determinant for specific protein-protein interactions.[3] For example, lipid groups can play an important role in increasing molecular hydrophobicity. This allows for the interaction of proteins with cellular membranes and protein domains.[4] In a dynamic role[clarification needed], lipidation can sequester a protein away from its substrate to inactivate the protein and then activate it by substrate presentation.
Overall, there are three main types of lipid-anchored proteins which include prenylated proteins, fatty acylated proteins and glycosylphosphatidylinositol-linked proteins (GPI).[2][5] A protein can have multiple lipid groups covalently attached to it, but[clarification needed] the site where the lipids bind to the protein depends both on the lipid group and protein.[2]
Prenylated proteins
[edit]
Prenylated proteins are proteins with covalently attached hydrophobic isoprene polymers (i.e. branched five-carbon hydrocarbon[6]) at cysteine residues of the protein.[2][3] More specifically, these isoprenoid groups, usually farnesyl (15-carbon) and geranylgeranyl (20-carbon) are attached to the protein via thioether linkages at cysteine residues near the C terminal of the protein.[3][4] This prenylation of lipid chains to proteins facilitate their interaction with the cell membrane.[1]

The prenylation motif “CaaX box” is the most common prenylation site in proteins, that is, the site where farnesyl or geranylgeranyl covalently attach.[2][3] In the CaaX box sequence, the C represents the cysteine that is prenylated, the A represents any aliphatic amino acid and the X determines the type of prenylation that will occur. If the X is an Ala, Met, Ser or Gln the protein will be farnesylated via the farnesyltransferase enzyme and if the X is a Leu then the protein will be geranylgeranylated via the geranylgeranyltransferase I enzyme.[3][4] Both of these enzymes are similar with each containing two subunits.[7]
Roles and function
[edit]
Prenylated proteins are particularly important for eukaryotic cell growth, differentiation and morphology.[7] Furthermore, protein prenylation is a reversible post-translational modification to the cell membrane. This dynamic interaction of prenylated proteins with the cell membrane is important for their signalling functions and is often deregulated in disease processes such as cancer.[8] More specifically, Ras is the protein that undergoes prenylation via farnesyltransferase and when it is switched on it can turn on genes involved in cell growth and differentiation. Thus overactiving Ras signalling can lead to cancer.[9] An understanding of these prenylated proteins and their mechanisms have been important for the drug development efforts in combating cancer.[10] Other prenylated proteins include members of the Rab and Rho families as well as lamins.[7]
Some important prenylation chains that are involved in the HMG-CoA reductase metabolic pathway[1] are geranylgeraniol, farnesol and dolichol. These isoprene polymers (e.g. geranyl pyrophosphate and farnesyl pyrophosphate) are involved in the condensations via enzymes such as prenyltransferase that eventually cyclizes to form cholesterol.[2]
Fatty acylated proteins
[edit]Fatty acylated proteins are proteins that have been post-translationally modified to include the covalent attachment of fatty acids at certain amino acid residues.[11][12] The most common fatty acids that are covalently attached to the protein are the saturated myristic (14-carbon) acid and palmitic acid (16-carbon). Proteins can be modified to contain either one or both of these fatty acids.[11]

N-myristoylation
[edit]N-myristoylation (i.e. attachment of myristic acid) is generally an irreversible protein modification that typically occurs during protein synthesis[11][13] in which the myrisitc acid is attached to the α-amino group of an N-terminal glycine residue through an amide linkage.[2][12] This reaction is facilitated by N-myristoyltransferase . These proteins usually begin with a Met-Gly sequence and with either a serine or threonine at position 5.[11] Proteins that have been myristoylated are involved in signal transduction cascade, protein-protein interactions and in mechanisms that regulate protein targeting and function.[13] An example in which the myristoylation of a protein is important is in apoptosis, programmed cell death. After the protein BH3 interacting-domain death agonist (Bid) has been myristoylated, it targets the protein to move to the mitochondrial membrane to release cytochrome c, which then ultimately leads to cell death.[14] Other proteins that are myristoylated and involved in the regulation of apoptosis are actin and gelsolin.
S-palmitoylation
[edit]
S-palmitoylation (i.e. attachment of palmitic acid) is a reversible protein modification in which a palmitic acid is attached to a specific cysteine residue via thioester linkage.[2][11] The term S-acylation can also be used when other medium and long fatty acids chains are also attached to palmitoylated proteins. No consensus sequence for protein palmitoylation has been identified.[11] Palmitoylated proteins are mainly found on the cytoplasmic side of the plasma membrane where they play a role in transmembrane signaling. The palmitoyl group can be removed by palmitoyl thioesterases. It is believed that this reverse palmitoylation may regulate the interaction of the protein with the membrane and thus have a role in signaling processes.[2] Furthermore, this allows for the regulation of protein subcellular localization, stability and trafficking.[15] An example in which palmitoylation of a protein plays a role in cell signaling pathways is in the clustering of proteins in the synapse. When the postsynaptic density protein 95 (PSD-95) is palmitoylated, it is restricted to the membrane and allows it to bind to and cluster ion channels in the postsynaptic membrane. Thus, palmitoylation can play a role in the regulation of neurotransmitter release.[16]
Palmitoylation mediates the affinity of a protein for lipid rafts and facilitates the clustering of proteins.[17] The clustering can increase the proximity of two molecules. Alternatively, clustering can sequester a protein away from a substrate. For example, palmitoylation of phospholipase D (PLD) sequesters the enzyme away from its substrate phosphatidylcholine. When cholesterol levels decrease or PIP2 levels increase the palmitate mediated localization is disrupted, the enzyme trafficks to PIP2 where it encounters its substrate and is active by substrate presentation.[18][19][20]
GPI proteins
[edit]
Glycosylphosphatidylinositol-anchored proteins (GPI-anchored proteins) are attached to a GPI complex molecular group via an amide linkage to the protein's C-terminal carboxyl group.[21] This GPI complex consists of several main components that are all interconnected: a phosphoethanolamine, a linear tetrasaccharide (composed of three mannose and a glucosaminyl) and a phosphatidylinositol.[22] The phosphatidylinositol group is glycosidically linked to the non-N-acetylated glucosamine of the tetrasaccharide. A phosphodiester bond is then formed between the mannose at the nonreducing end (of the tetrasaccaride) and the phosphoethanolamine. The phosphoethanolamine is then amide linked to the C-terminal of the carboxyl group of the respective protein.[2] The GPI attachment occurs through the action of GPI-transamidase complex.[22] The fatty acid chains of the phosphatidylinositol are inserted into the membrane and thus are what anchor the protein to the membrane.[23] These proteins are only located on the exterior surface of the plasma membrane.[2]
Roles and function
[edit]The sugar residues in the tetrasaccaride and the fatty acid residues in the phosphatidylinositol group vary depending on the protein.[2] This great diversity is what allows the GPI proteins to have a wide range of functions including acting as hydrolytic enzymes, adhesion molecule, receptors, protease inhibitor and complement regulatory proteins.[24] Furthermore, GPI proteins play an important in embryogenesis, development, neurogenesis, the immune system and fertilization.[21] More specifically, the GPI protein IZUMO1R (also named JUNO after the Roman goddess of fertility) on the egg plasma has an essential role in sperm-egg fusion. Releasing the IZUMO1R (JUNO) GPI protein from the egg plasma membrane does not allow for sperm to fuse with the egg and it is suggested that this mechanism may contribute to the polyspermy block at the plasma membrane in eggs.[25] Other roles that GPI modification allows for is in the association with membrane microdomains, transient homodimerization or in apical sorting in polarized cells.[21]
References
[edit]- ^ a b c Gerald Karp (2009). Cell and Molecular Biology: Concepts and Experiments. John Wiley and Sons. pp. 128–. ISBN 978-0-470-48337-4. Retrieved 13 November 2010.
- ^ a b c d e f g h i j k l m Voet D, Voet JG, Pratt CW (2013). Fundamentals of Biochemistry: Life at the Molecular Level (4th ed.). John Wiley & Sons, Inc. p. 263. ISBN 978-0470-54784-7.
- ^ a b c d e Casey PJ, Seabra MC (March 1996). "Protein prenyltransferases". The Journal of Biological Chemistry. 271 (10): 5289–92. doi:10.1074/jbc.271.10.5289. PMID 8621375.
- ^ a b c Novelli G, D'Apice MR (September 2012). "Protein farnesylation and disease". Journal of Inherited Metabolic Disease. 35 (5): 917–26. doi:10.1007/s10545-011-9445-y. PMID 22307208. S2CID 11555502.
- ^ Ferguson MA (August 1991). "Lipid anchors on membrane proteins". Current Opinion in Structural Biology. 1 (4): 522–9. doi:10.1016/s0959-440x(05)80072-7.
- ^ isoprene (2003). "Miller-Keane Encyclopedia and Dictionary of Medicine, Nursing, and Allied Health, Seventh Ed". Retrieved 28 November 2015.
- ^ a b c Lane KT, Beese LS (April 2006). "Thematic review series: lipid posttranslational modifications. Structural biology of protein farnesyltransferase and geranylgeranyltransferase type I". Journal of Lipid Research. 47 (4): 681–99. doi:10.1194/jlr.R600002-JLR200. PMID 16477080.
- ^ Stein V, Kubala MH, Steen J, Grimmond SM, Alexandrov K (2015-01-01). "Towards the systematic mapping and engineering of the protein prenylation machinery in Saccharomyces cerevisiae". PLOS ONE. 10 (3): e0120716. Bibcode:2015PLoSO..1020716S. doi:10.1371/journal.pone.0120716. PMC 4358939. PMID 25768003.
- ^ Goodsell DS (1999-01-01). "The molecular perspective: the ras oncogene". The Oncologist. 4 (3): 263–4. doi:10.1634/theoncologist.4-3-263. PMID 10394594.
- ^ Reuter CW, Morgan MA, Bergmann L (September 2000). "Targeting the Ras signaling pathway: a rational, mechanism-based treatment for hematologic malignancies?". Blood. 96 (5): 1655–69. doi:10.1182/blood.V96.5.1655. PMID 10961860.
- ^ a b c d e f Resh MD (November 2006). "Trafficking and signaling by fatty-acylated and prenylated proteins". Nature Chemical Biology. 2 (11): 584–90. doi:10.1038/nchembio834. PMID 17051234. S2CID 9734759.
- ^ a b Wilson JP, Raghavan AS, Yang YY, Charron G, Hang HC (March 2011). "Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants". Molecular & Cellular Proteomics. 10 (3): M110.001198. doi:10.1074/mcp.M110.001198. PMC 3047146. PMID 21076176.
- ^ a b Farazi TA, Waksman G, Gordon JI (October 2001). "The biology and enzymology of protein N-myristoylation". The Journal of Biological Chemistry. 276 (43): 39501–4. doi:10.1074/jbc.R100042200. PMID 11527981.
- ^ Martin DD, Beauchamp E, Berthiaume LG (January 2011). "Post-translational myristoylation: Fat matters in cellular life and death". Biochimie. Bioactive Lipids, Nutrition and Health. 93 (1): 18–31. doi:10.1016/j.biochi.2010.10.018. PMID 21056615.
- ^ Aicart-Ramos C, Valero RA, Rodriguez-Crespo I (December 2011). "Protein palmitoylation and subcellular trafficking". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1808 (12): 2981–94. doi:10.1016/j.bbamem.2011.07.009. PMID 21819967.
- ^ Dityatev, Alexander (2006). El-Husseini, Alaa (ed.). Molecular Mechanisms of Synaptogenesis. New York: Springer. pp. 72–75.
- ^ Levental, I.; Lingwood, D.; Grzybek, M.; Coskun, U.; Simons, K. (3 December 2010). "Palmitoylation regulates raft affinity for the majority of integral raft proteins". Proceedings of the National Academy of Sciences. 107 (51): 22050–22054. Bibcode:2010PNAS..10722050L. doi:10.1073/pnas.1016184107. PMC 3009825. PMID 21131568.
- ^ Petersen, EN; Chung, HW; Nayebosadri, A; Hansen, SB (15 December 2016). "Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D." Nature Communications. 7: 13873. Bibcode:2016NatCo...713873P. doi:10.1038/ncomms13873. PMC 5171650. PMID 27976674.
- ^ Robinson, CV; Rohacs, T; Hansen, SB (September 2019). "Tools for Understanding Nanoscale Lipid Regulation of Ion Channels". Trends in Biochemical Sciences. 44 (9): 795–806. doi:10.1016/j.tibs.2019.04.001. PMC 6729126. PMID 31060927.
- ^ Petersen, EN; Pavel, MA; Wang, H; Hansen, SB (28 October 2019). "Disruption of palmitate-mediated localization; a shared pathway of force and anesthetic activation of TREK-1 channels". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1862 (1): 183091. doi:10.1016/j.bbamem.2019.183091. PMC 6907892. PMID 31672538.
- ^ a b c Kinoshita T, Fujita M (January 2016). "Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling". Journal of Lipid Research. 57 (1): 6–24. doi:10.1194/jlr.R063313. PMC 4689344. PMID 26563290.
- ^ a b Ikezawa, Hiroh (2002-01-01). "Glycosylphosphatidylinositol (GPI)-Anchored Proteins". Biological and Pharmaceutical Bulletin. 25 (4): 409–417. doi:10.1248/bpb.25.409. PMID 11995915.
- ^ Kinoshita T, Fujita M (January 2016). "Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling". Journal of Lipid Research. 57 (1): 6–24. doi:10.1194/jlr.R063313. PMC 4689344. PMID 26563290.
- ^ Kinoshita T (2014). "Biosynthesis and deficiencies of glycosylphosphatidylinositol". Proceedings of the Japan Academy. Series B, Physical and Biological Sciences. 90 (4): 130–43. Bibcode:2014PJAB...90..130K. doi:10.2183/pjab.90.130. PMC 4055706. PMID 24727937.
- ^ Coonrod SA, Naaby-Hansen S, Shetty J, Shibahara H, Chen M, White JM, Herr JC (March 1999). "Treatment of mouse oocytes with PI-PLC releases 70-kDa (pI 5) and 35- to 45-kDa (pI 5.5) protein clusters from the egg surface and inhibits sperm-oolemma binding and fusion". Developmental Biology. 207 (2): 334–49. doi:10.1006/dbio.1998.9161. PMID 10068467.
External links
[edit] Media related to Lipid-anchored protein at Wikimedia Commons
Media related to Lipid-anchored protein at Wikimedia Commons
